A New Era of
Neurodegenerative
Drug Development
We're developing novel therapeutics to enhance lysosome function and increase autophagy. By enhancing the body’s natural ability to deliver and remove dysfunctional proteins and machinery, our therapeutics could slow disease progression and improve patients’ day-to-day lives.

Our lead program is in pre-clinical development with line-of-sight to initiate phase 1 studies
The company’s therapeutic platform is uniquely positioned to discover and develop novel small molecule drugs that can both increase autophagy to more rapidly clear toxic proteins and dysfunctional machinery, as well as reduce the production of neurotoxic proteins. Our lead program from Libra’s Lysofunction Platform targets TRPML1, a regulator of autophagy and lysosome function.
-
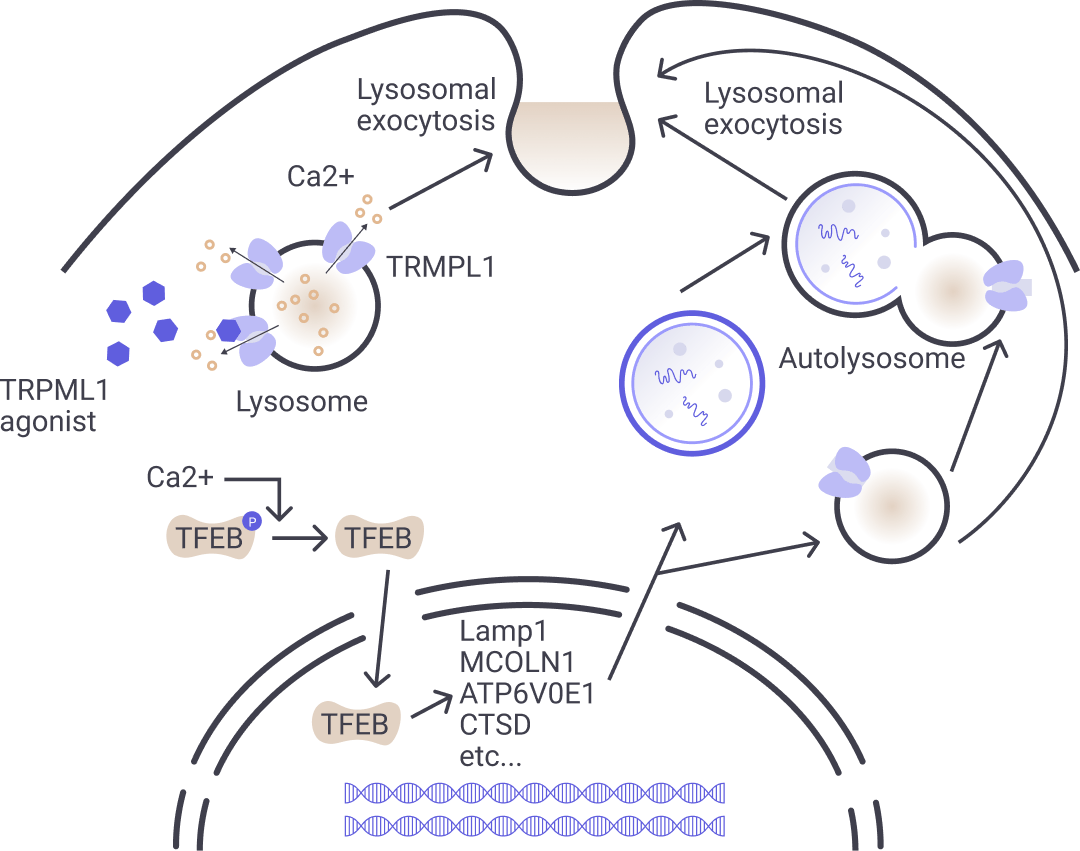
Enhancing lysosomal function via TRPML1 activation
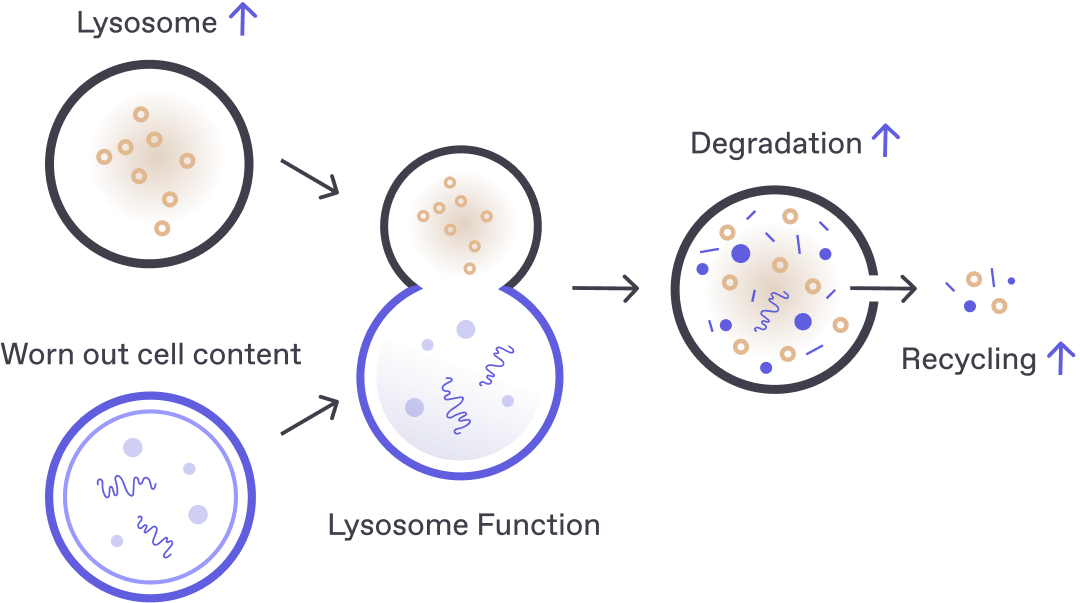
Protein accumulation and/or defective cellular clearance are impaired in most neurodegenerative diseases, including amyotrophic lateral Sclerosis, Alzheimer’s disease, and Parkinson’s disease. Regulating the buildup of “trash" within the cell by maintaining the balance between protein accumulation and cellular clearance is essential for healthy neurons.
Libra Therapeutics’ approach focuses on pharmacological activation of the lysosomal calcium ion channel, TRPML1, which leads to increased macroautophagy, TFEB-mediated lysosomal biogenesis and membrane repair. We believe this mechanism can both reduce trash accumulation and augment its clearance within a cell to help return balance to a healthy state.
-
Libra’s Lysofunction Platform
Lysosomal dysfunction has been implicated in the pathology of several major disease areas including inflammation, neurodegeneration, and cancer. Libra Therapeutics has established a screening platform, which identifies compounds that enhance autophagy and lysosomal biogenesis both in cells and in animals. The platform is both time and cost effective and can be used to evaluate a compound’s or target’s potential to enhance autophagy and increase lysosome function. As more compounds and targets are identified which have a significant effect on the function of the lysosome, they will be valuable tools to further explore a role for the lysosome in the pathology of disease and ultimately help to identify novel and safe therapeutics.
-
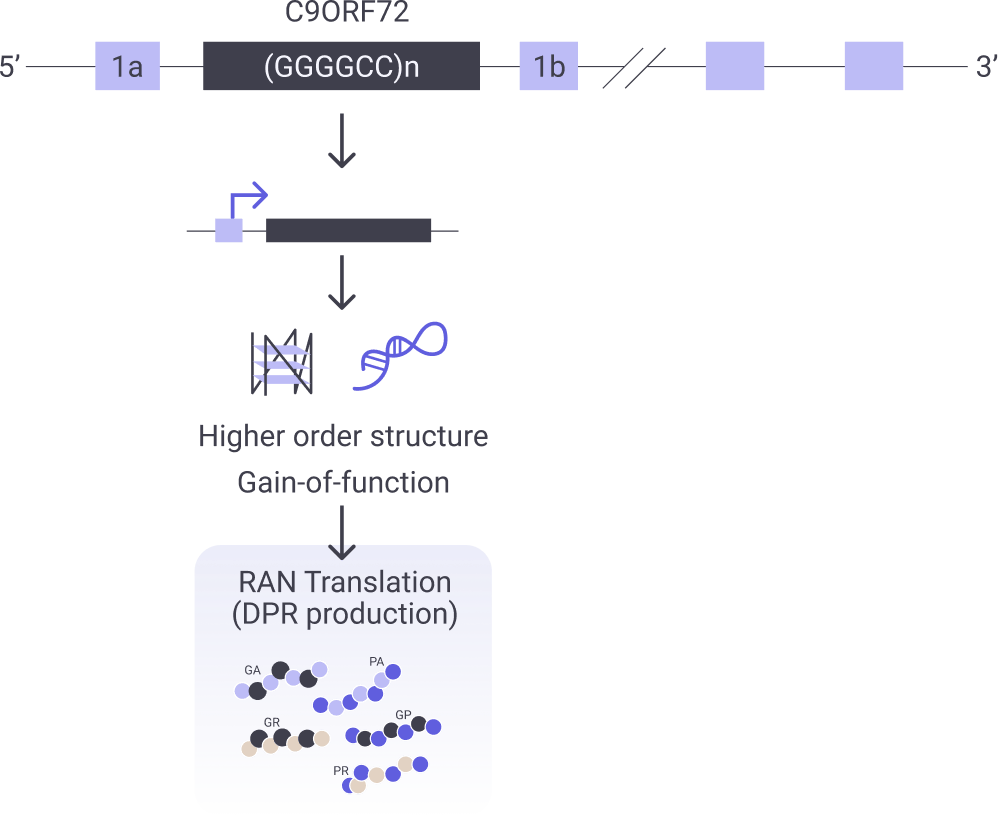
RAN-Translation Inhibition
Nucleotide repeat expansions produce dipeptide-repeat-containing proteins (DPRs) which cause multiple neurodegenerative disorders, including amyotrophic lateral sclerosis (ALS), Huntington’s disease, frontotemporal dementia (FTD), fragile X syndrome and Deuchene’s muscular dystrophy to name a few. DPRs induce toxicity through a number of mechanisms, including altered ribosomal biogenesis, impaired nucleocytoplasmic transport, shifts in RNA metabolism, protein sequestration, and impaired protein quality control pathways. C9orf72 is the most common known inherited cause of both ALS and FTD and repeat expansion of the C9orf72 gene leads to the production of toxic C9orf72 DPRs. Libra Therapeutics’ approach is to identify novel chemical matter, acting on several processes within the pathway, that selectively inhibit the translation of repeat expansions into toxic protein species.
-
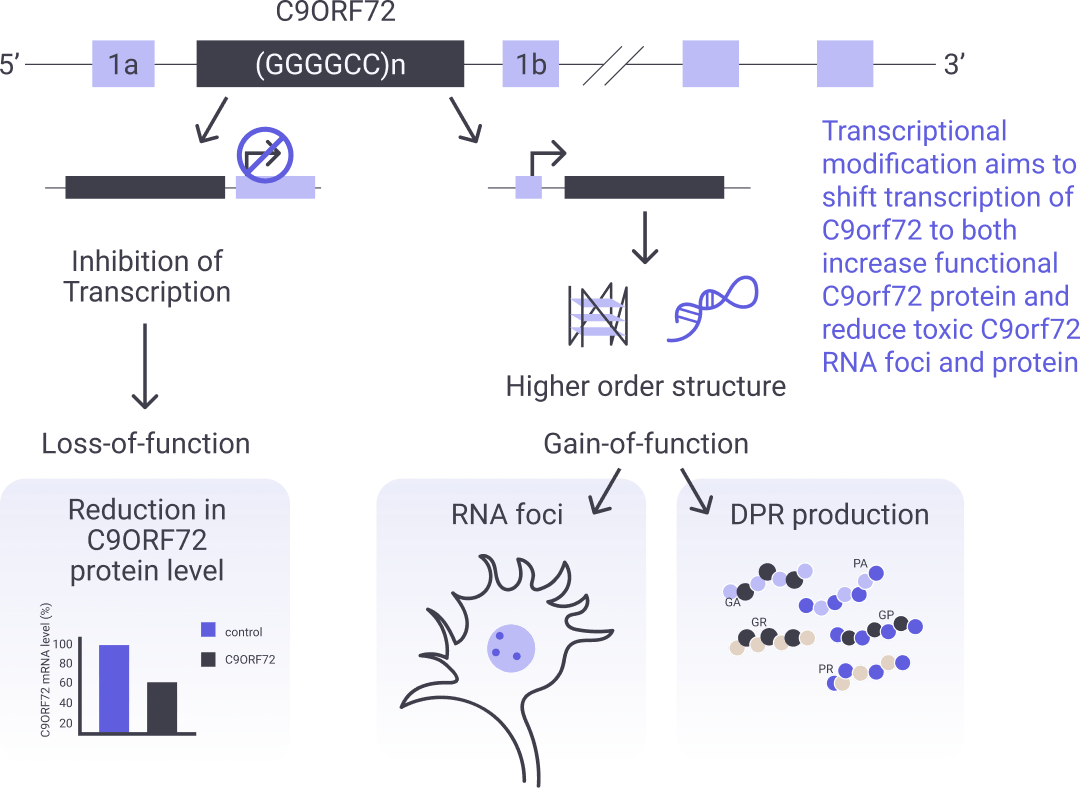
Modulation of the C9orf72 function
C9orf72 is a gene locus whose products have been shown to regulate lysosomal function, autophagy and inflammation. The C9orf72 non-coding repeat expansion is the most common genetic link to frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). This C9orf72 non-coding repeat expansion is associated with a loss of function, resulting from the disruption of normal gene transcription, and a gain of toxicity, resulting from the aberrant production of dipeptide repeats from the non-coding repeat expansion. Our two CP9orf72 programs target these two core disease processes. Our RAN translation inhibition program seeks to identify small molecules, which block the RAN-mediated production of dipeptide repeats, whereas our small molecule C9orf72 transcriptional modification program seeks to boost normal gene transcription.